the outline of the beast.
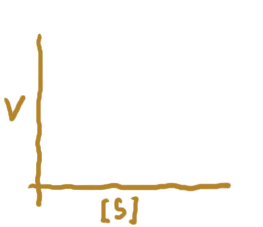
So on last page, we figured out that we are trying to produce a graph where the x-axis is the amount of the substrate and the y-axis is the velocity of the reaction. Here are the axes:
| If you turn on javascript, this becomes a rollover |
Instead of "amount of substrate", I put [S]. S is an abbreviation, and the square brackets, as you know, mean that we are dealing with concentration -- that is, how much substrate there is PER VOLUME of space. One unit of substrate in a testtube is very different from the same amount of substrate in a single cell, or the same amount of substrate in the entire body.

Let's think about what you know about the chocolatase enzyme. Imagine the following situation: a huge room, with bags of chocolate chips laying here and there on the floor, and a single chocolatase enzyme oozing around. How fast could the enzyme turn the bags of chocolate chips into piles of mush?
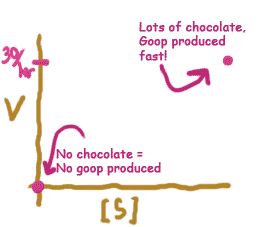
Well, if there were lots and lots of bags of chocolate chips, the enzyme would need to spend any time looking for a new bag. Does that mean that the bags make it converted into mush at an infinitely fast rate? No, it still takes a certain finite amount of time for the enzyme to process a single bag. Let's say an enzyme can go through a single bag in two minutes. That means that, no matter how many bags of chocolate chips there are, goop will never be produced faster than .5 piles per minute.
On the other end of the scale, what if there are no bags of chocolate chips? In other words,[S] = 0? Pretty obviously, no piles of mush we get produced.
Now we know what two parts of this graph should look like. Try sketching the axes yourself, on a piece of scratch paper, and filling in our predictions for lots of substrate, and no substrate. When you're done, click on the button below.
| If you turn on javascript, this becomes a rollover |
Copyright University of Maryland, 2007
You may link to this site for educational purposes.
Please do not copy without permission
requests/questions/feedback email: mathbench@umd.edu