Light scatter (using spec)
 Direct count is a straightforward way to figure out how many meningicocci Frank is harboring, but it is a bit labor intensive. Wouldn't it be nice to get a machine to do the counting for us?
Direct count is a straightforward way to figure out how many meningicocci Frank is harboring, but it is a bit labor intensive. Wouldn't it be nice to get a machine to do the counting for us?
One way we can do this is to use a photospectometer. A spec doesn't exactly count the cocci, but it does measure how much they interfere with a light beam, and based on that, we can use a calibration to decide how many cocci we've got. Then we can do the scaling up trick to get the total population size.
This is a great method if you already have a calibration curve -- perhaps you work in a lab that has been studying this critter for years... Otherwise, you may need to spend a couple of days doing the calibration (growing the cultures, taking the spec readings, comparing them to counts under the microscope... that sort of thing).
Here's an example of a calibration curve:
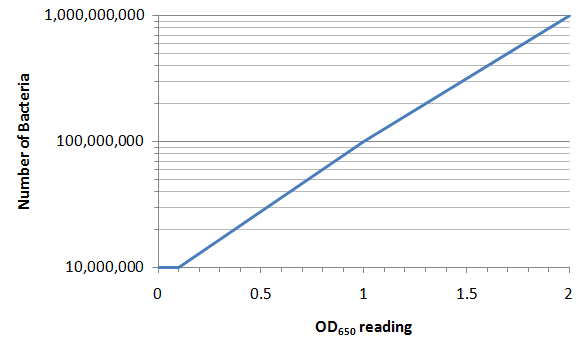
For example, if you prepared a sample and got a spec reading of 0.5, you would find 0.5 on the x-axis and read the bacterial count off the y-axis -- about 26 million
How about a spec reading of 0.9? (Be careful with the log scale for bacteria!!)
If your sample had about 12 million bacteria, what OD reading would you expect?
Copyright University of Maryland, 2007
You may link to this site for educational purposes.
Please do not copy without permission
requests/questions/feedback email: mathbench@umd.edu